Introduction
This guide shows where to find the image acquisition and image data information from raw data. These two components are part of the complete metadata of a study . For further reading REMBI: Recommended Metadata for biological images-enabling reuse of microscopy data in Biology
-
-
Open FiJI
-
In Tab File, select Import and Bio-format
-
Select the image to open.
-
A new window will appear.
-
In the metadata viewing part. Check to activate Display metadata and Display metadata OME-XML metadata
-
Two windows with the metadata information will appear together with the image
-
-
-
Open the file on LAS application
-
Right-click on the image you want to know its metadata
-
Select Properties
-
A new window will appear showing the metadata.
-
Leica data format is .lif
-
-
-
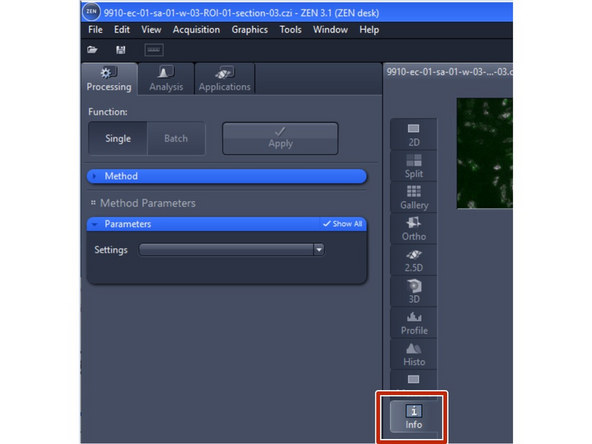
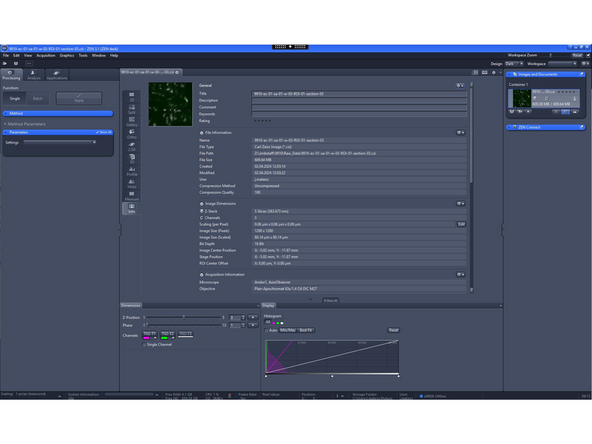
Open the data with ZEN sofware
-
Click on the info tab to see the metadata values
-
Zeiss data format is .czi
-
-
-
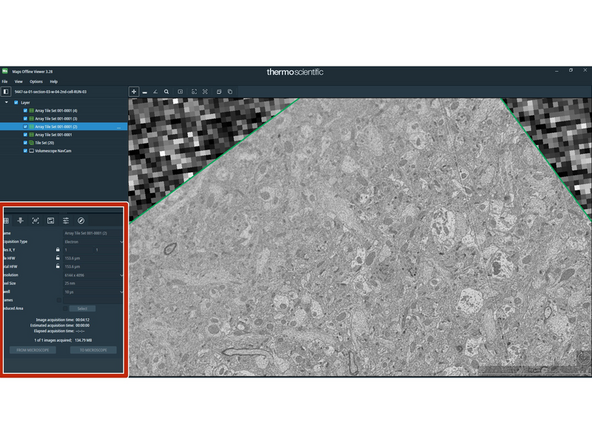
Open the data with MAPS and on the lower-left corner you can read some metadata parameters.
-
-
-
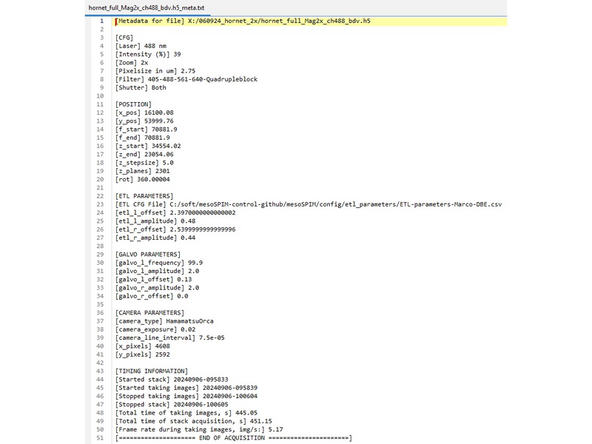
MesoSPIM saves the data in h5 format and the metadata can be found in an associated .txt file. Open it with any text editor.
-
MesoSPIM data format is .h5
-
-
-
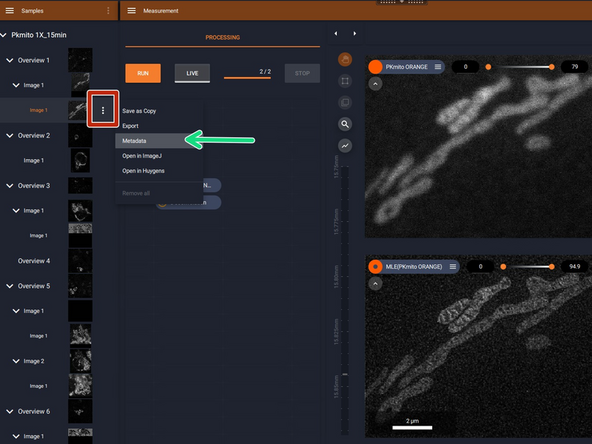
Open the data with Abberior Lightbox software.
-
Click on the three points icon on the right side of each image.
-
Select metadata.
-
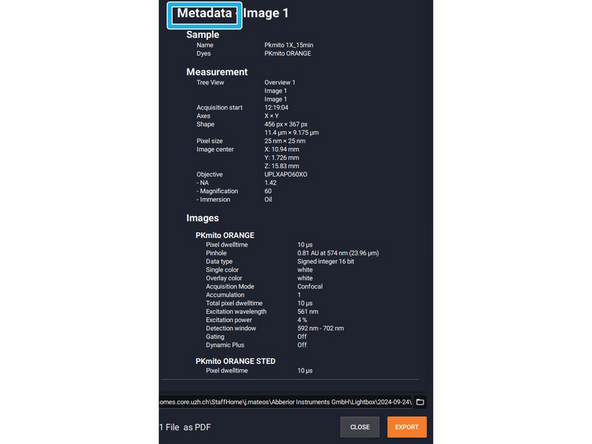
A new window will appear with the metadata information.
-
Abberior data format is .obf
-
-
-
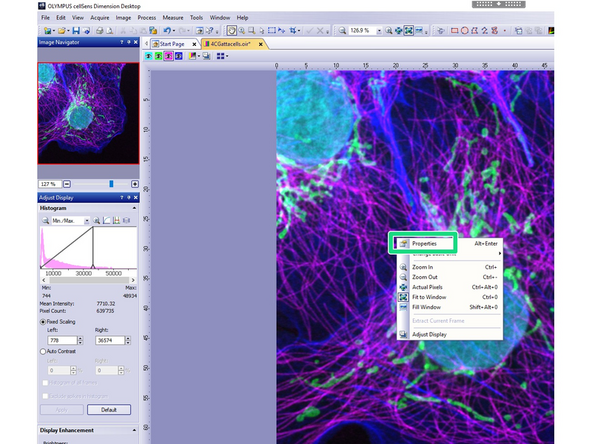
Open the data with CellSens Dimension Desktop.
-
Right-Click on the image and from the appearing window, select properties.
-
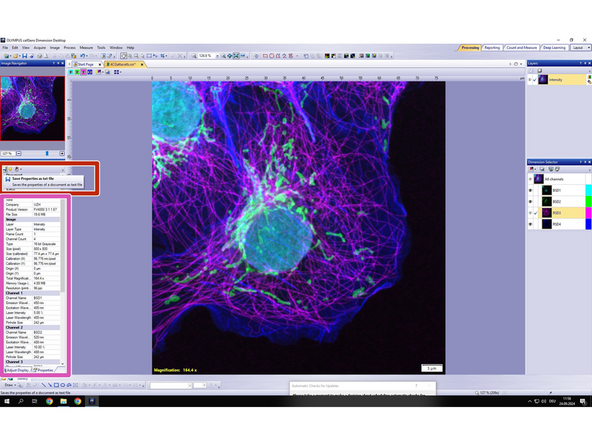
A new window with the metadata will appear.
-
This metadata can be exported as .txt file by clicking on the "save properties as txt file" icon.
-
Evident data format is .oir
-
-
-
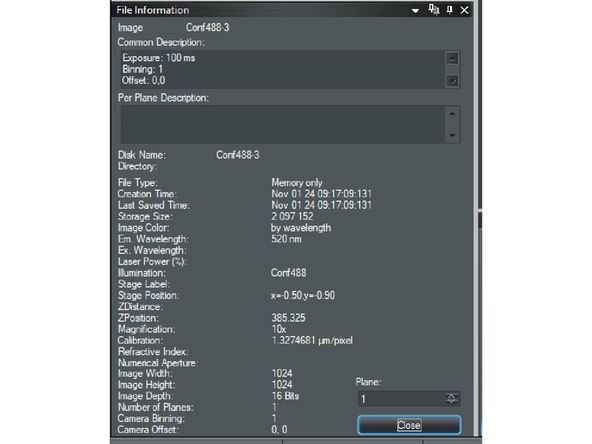
Open the file with VisiView software.
-
In the Main menu select: "File" and from the drop-down menu select: "File Info".
-
A window will appear with the image metadata.
-
Data can be saved in Visitron data format .stk
-
or in ome.tf2 Format OME Big-Tiff (recommended)
-