-
-
Collect data using SIM mode with the same imaging parameters as your sample.
-
Acquire a z-Stack of TetraSpeckTM microspheres spread onto #1.5 coverslips and embedded in a hardening mounting media of your choice with a accurate step size in the relevant colors with SIM (identical settings as your experiment) and process the data with the ‘SIM’ processing tool.
-
Alternatively, use the microspheres slide from Carl Zeiss Microscopy GmbH.
-
This acquired stack will be used to calibrate the color offset of the system. Offsets will be stored in a template that later on can be used to correct the experimental data.
-
Run the Structured Illumination processing as described in the guide.
-
Calibrated color offsets are specific for the objective and filters. Therefore, a new calibration is required when objectives and filters are exchanged.
-
-
-
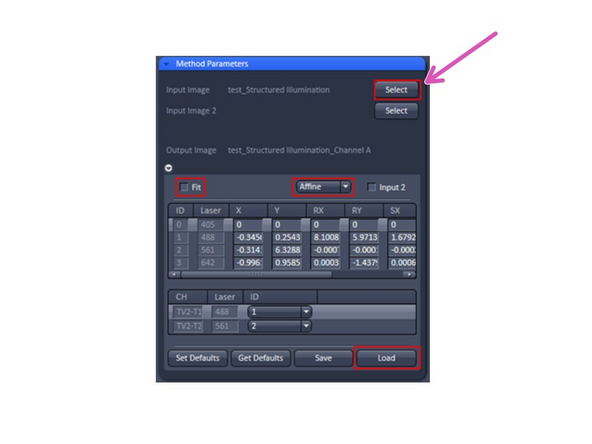
In the Processing tab select the Channel Alignment tool from the list of Methods.
-
In the Method Parameters tab select the file that you obtained from the Structured Illumination processing of your bead stack by opening the file in ZEN and clicking Select. The filename of the selected dataset will appear next to the button.
-
In the Channel Alignment options make sure Fit is checked and select Affine, Lateral or Parabolic from the dropdown menu.
-
Lateral is used to correct for linear lateral and axial drifts, that is in all directions. Linear alignments are performed in the x, y and z direction.
-
Affine is used to correct for linear lateral (X, Y) and axial (Z) drifts, rotational offsets in the image plane in x and y (RX, RY) as well as a 1st order stretching in the image plane in x and y (SX, SY).
-
Press Apply near the top of the screen to start the processing (takes some time to run).
-
In the resulting image all colour channels should be properly aligned now. Save the transformation table on the left as a ".bin" file by clicking the Save-button.
-
-
-
In the Processing tab select the Channel Alignment tool from the list of Methods.
-
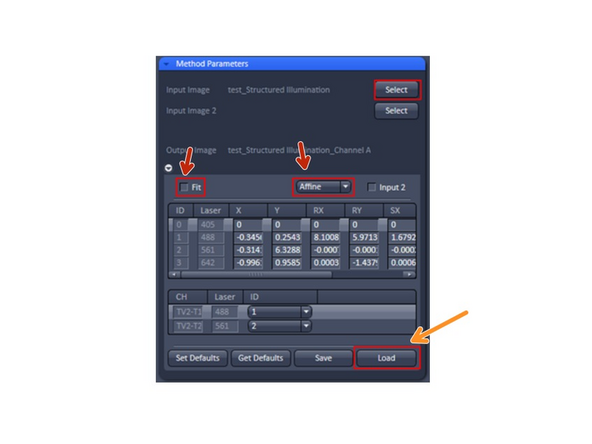
In the Method Parameters tab select the Structured Illumination output image of your sample that you want to correct for chromatic aberrations by opening it in ZEN and clicking Select. The filename of the selected dataset will appear next to the button.
-
-
-
Unselect the ‘Fit’ check box
-
Then click on the Load-button under the empty transformation table, select the bin file you saved earlier and press Open. Now the table should display the values for the colour shifts between channels.
-
.bin file is used to compensate for any chromatic aberrations and offsets between channels in SIM.
-
In the table below make sure the wavelength channel IDs correspond to the wavelengths of the channels in the transformation table above.
-
Press Apply to apply the transformation table to your sample image.
-
Save the resulting file in .czi format. ZEN will automatically append “Channel Alignment” to the output filename.
-
Refractive index mismatches don't affect the correction template as the beads are on the coverslip. If the sample is fixed to the coverslip, the correction is reliable regardless of embedding differences. However, if fixed on the slide, mismatches may impact accuracy, requiring beads to be embedded in the same medium as the sample.
-
Almost done!
Finish Line